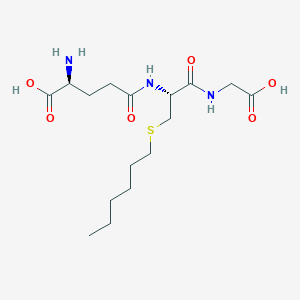
Details of Metabolite
| Full List of Protein(s) Regulating This Metabolite | ||||||
|---|---|---|---|---|---|---|
| Transcriptional coactivator (TC) | ||||||
| PPAR-gamma coactivator 1-alpha (PPARGC1A) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[1] | ||||
| Introduced Variation | Knockout of Ppargc1a | |||||
| Induced Change | s-Hexylglutathione concentration: decrease | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Congestive heart failure [ICD-11: BD10] | |||||
| Details | It is reported that knockout of Ppargc1a leads to the decrease of s-hexylglutathione levels compared with control group. | |||||
| Transferases (EC 2) | ||||||
| Arylamine N-acetyltransferase 1 (NAT1) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[2] | ||||
| Introduced Variation | Knockout of Nat1 | |||||
| Induced Change | s-Hexylglutathione concentration: decrease | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Metabolic liver disease [ICD-11: 5C90] | |||||
| Details | It is reported that knockout of NAT leads to the decrease of s-hexylglutathione levels compared with control group. | |||||
| Arylamine N-acetyltransferase 2 (NAT2) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[2] | ||||
| Introduced Variation | Knockout of Nat2 | |||||
| Induced Change | s-Hexylglutathione concentration: decrease | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Metabolic liver disease [ICD-11: 5C90] | |||||
| Details | It is reported that knockout of NAT leads to the decrease of s-hexylglutathione levels compared with control group. | |||||
If you find any error in data or bug in web service, please kindly report it to Dr. Zhang and Dr. Mou.