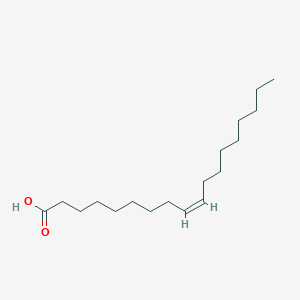
Details of Metabolite
| Full List of Protein(s) Regulated by This Metabolite | ||||||
|---|---|---|---|---|---|---|
| Cytokine receptor (CytR) | ||||||
| Apoptosis mediating antigen FAS (CD95) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[1] | ||||
| Introduced Variation | Oleic acid addition (24 hours) | |||||
| Induced Change | FAS protein expression levels: decrease | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Hepatocellular carcinoma [ICD-11: 2C12] | |||||
| Details | It is reported that oleic acid addition causes the decrease of FAS protein expression compared with control group. | |||||
| GPCR rhodopsin (GPCR-1) | ||||||
| G-protein coupled receptor 120 (GPR120) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[2] | ||||
| Introduced Variation | Oleic acid addition (40.08 hours) | |||||
| Induced Change | FFAR4 protein activity levels: increase | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Healthy individual | |||||
| Details | It is reported that oleic acid addition causes the increase of FFAR4 protein activity compared with control group. | |||||
| Full List of Protein(s) Regulating This Metabolite | ||||||
|---|---|---|---|---|---|---|
| Hydrolases (EC 3) | ||||||
| Group 3 secretory phospholipase A2 (PLA2G3) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[3] | ||||
| Introduced Variation | Knockout of Pla2g3 | |||||
| Induced Change | Oleic acid concentration: increase | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Colorectal cancer [ICD-11: 2B91] | |||||
| Details | It is reported that knockout of Pla2g3 leads to the increase of oleic acid levels compared with control group. | |||||
| Sulfatase sulf-1 (SULF1) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[4] | ||||
| Introduced Variation | Knockdown (shRNA) of SULF1 | |||||
| Induced Change | Oleic acid concentration: increase (FC = 2.22 / 2.70) | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Ovarian cancer [ICD-11: 2C73] | |||||
| Details | It is reported that knockdown of SULF1 leads to the increase of oleic acid levels compared with control group. | |||||
| Transcription factor (TF) | ||||||
| Hypoxia-inducible factor 1-alpha (HIF1A) | Click to Show/Hide the Full List of Regulating Pair(s): 1 Pair(s) | |||||
| Detailed Information |
Protein Info
 click to show the details of this protein click to show the details of this protein
|
|||||
| Regulating Pair |
Experim Info
 click to show the details of experiment for validating this pair click to show the details of experiment for validating this pair
|
[5] | ||||
| Introduced Variation | Knockout of HIF1A | |||||
| Induced Change | Oleic acid concentration: increase | |||||
| Summary | Introduced Variation
|
|||||
| Disease Status | Colorectal cancer [ICD-11: 2B91] | |||||
| Details | It is reported that knockout of HIF1A leads to the increase of oleic acid levels compared with control group. | |||||
If you find any error in data or bug in web service, please kindly report it to Dr. Zhang and Dr. Mou.