| Synonyms |
Click to Show/Hide Synonyms of This Metabolite
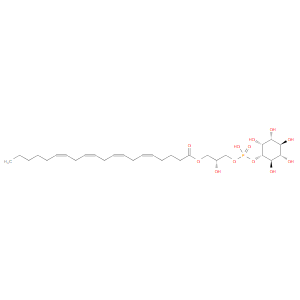
1-(5Z,8Z,11Z,14Z-Eicosatetraenoyl)-sn-glycero-3-phospho-(1'-myo-inositol), '1-Arachidonoylglycerophosphoinositol; 1-Arachidonoyl-lysophosphatidylinositol; 1-Arachidonoyl-sn-glycero-3-phospho-D-myo-inositol; GPI(20:4(5Z,8Z,11Z,14Z)); GPI(20:4(5Z,8Z,11Z,14Z)/0:0); GPI(20:4); GPI(20:4W6); GPI(20:4W6/0:0); GPI(20:4n6); GPI(20:4n6/0:0); LPI(20:4(5Z,8Z,11Z,14Z)); LPI(20:4(5Z,8Z,11Z,14Z)/0:0); LPI(20:4); LPI(20:4W6); LPI(20:4W6/0:0); LPI(20:4n6); LPI(20:4n6/0:0); LysoPI(20:4(5Z,8Z,11Z,14Z)); LysoPI(20:4(5Z,8Z,11Z,14Z)/0:0); LysoPI(20:4); LysoPI(20:4W6); LysoPI(20:4W6/0:0); LysoPI(20:4n6); LysoPI(20:4n6/0:0); Lysophosphatidylinositol(20:4(5Z,8Z,11Z,14Z)); Lysophosphatidylinositol(20:4(5Z,8Z,11Z,14Z)/0:0); Lysophosphatidylinositol(20:4); Lysophosphatidylinositol(20:4W6); Lysophosphatidylinositol(20:4W6/0:0); Lysophosphatidylinositol(20:4n6); Lysophosphatidylinositol(20:4n6/0:0); PI(20:4(5Z,8Z,11Z,14Z)/0:0)
|
 click to show the details of this protein
click to show the details of this protein
 click to show the details of experiment for validating this pair
click to show the details of experiment for validating this pair