| General Information of MET (ID: META00435) |
| Name |
NADH
|
| Synonyms |
Click to Show/Hide Synonyms of This Metabolite
1,4-DIHYDRONICOTINAMIDE adenine dinucleotide; Adenine dinucleotide, dihydronicotinamide; Coenzyme I; DPN; DPNH; Dihydrocodehydrogenase I; Dihydrocozymase; Dihydronicotinamide adenine dinucleotide; Dihydronicotinamide mononucleotide; Dinucleotide, dihydronicotinamide adenine; Dinucleotide, nicotinamide-adenine; Diphosphopyridine nucleotide; ENADA; NAD; NADH2; Nadide; Nicotinamide adenine dinucleotide; Nicotinamide adenine dinucleotide (reduced); Nicotinamide-adenine dinucleotide; Nucleotide, diphosphopyridine; Reduced codehydrogenase I; Reduced diphosphopyridine nucleotide; Reduced nicotinamide adenine dinucleotide; Reduced nicotinamide adenine diphosphate; Reduced nicotinamide-adenine dinucleotide; b-DPNH; b-NADH; beta-DPNH; beta-NADH
|
| Source |
Endogenous;Escherichia Coli Metabolite;Yeast Metabolite;Food;Drug;Microbial
|
| Structure Type |
(5'->5')-dinucleotides (Click to Show/Hide the Complete Structure Type Hierarchy)
Nucleosides, nucleotides, and analogues
(5'->5')-dinucleotides
|
| PubChem CID |
|
| HMDB ID |
|
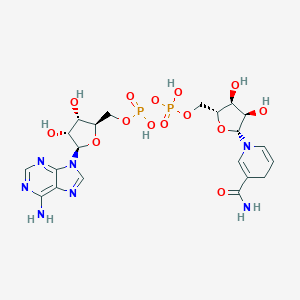
| Formula |
C21H29N7O14P2
|
| Structure |
<iframe style="width: 300px; height: 300px;" frameborder="0" src="https://embed.molview.org/v1/?mode=balls&cid=439153"></iframe>
|
 |
|
3D MOL
|
2D MOL
|
|
Click to Show/Hide the Molecular/Functional Data (External Links/Property/Function) of This Metabolite
|
| KEGG ID |
|
| DrugBank ID |
|
| ChEBI ID |
|
| FooDB ID |
|
| ChemSpider ID |
|
| METLIN ID |
|
| Physicochemical Properties |
Molecular Weight |
665.4 |
Topological Polar Surface Area |
318 |
| XlogP |
-5.7 |
Complexity |
1230 |
| Heavy Atom Count |
44 |
Rotatable Bond Count |
11 |
| Hydrogen Bond Donor Count |
8 |
Hydrogen Bond Acceptor Count |
19 |
| Function |
NADH is the reduced form of NAD+, and NAD+ is the oxidized form of NADH. NAD (or nicotinamide adenine dinucleotide) is used extensively in glycolysis and the citric acid cycle of cellular respiration. The reducing potential stored in NADH can be either converted into ATP through the electron transport chain or used for anabolic metabolism. ATP "energy" is necessary for an organism to live. Green plants obtain ATP through photosynthesis, while other organisms obtain it via cellular respiration. NAD is a coenzyme composed of ribosylnicotinamide 5'-diphosphate coupled to adenosine 5'-phosphate by a pyrophosphate linkage. It is found widely in nature and is involved in numerous enzymatic reactions in which it serves as an electron carrier by being alternately oxidized (NAD+) and reduced (NADH). NADP is formed through the addition of a phosphate group to the 2' position of the adenosyl nucleotide through an ester linkage (Dorland, 27th ed).
|
|
Regulatory Network
|
|
|
|
|
|
|
|
|
 click to show the details of this protein
click to show the details of this protein
 click to show the details of experiment for validating this pair
click to show the details of experiment for validating this pair

