| Synonyms |
Click to Show/Hide Synonyms of This Metabolite
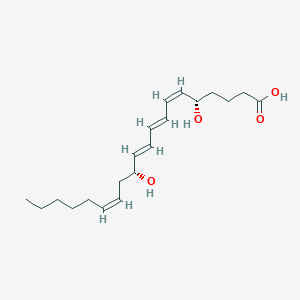
(+)-Leukotriene b4; (5S,12R)-Leukotriene b4; (5S,12R,6Z,8E,10E,14Z)-5,12-Dihydroxy-6,8,10,14-eicosatetraenoate; (5S,12R,6Z,8E,10E,14Z)-5,12-Dihydroxy-6,8,10,14-eicosatetraenoic acid; (5S,6Z,8E,10E,12R,14Z)-5,12-Dihydroxyeicosa-6,8,10,14-tetraenoate; (5S,6Z,8E,10E,12R,14Z)-5,12-Dihydroxyeicosa-6,8,10,14-tetraenoic acid; (6Z,8E,10E,14Z)-(5S,12R)-5,12-Dihydroxyeicosa-6,8,10,14-tetraenoate; (6Z,8E,10E,14Z)-(5S,12R)-5,12-Dihydroxyeicosa-6,8,10,14-tetraenoic acid; (6Z,8E,10E,14Z)-(5S,12R)-5,12-Dihydroxyicosa-6,8,10,14-tetraenoate; (6Z,8E,10E,14Z)-(5S,12R)-5,12-Dihydroxyicosa-6,8,10,14-tetraenoic acid; (S-(R*,s*-(e,Z,e,Z)))-5,12-dihydroxy-6,8,10,14-eicosatetraenoate; (S-(R*,s*-(e,Z,e,Z)))-5,12-dihydroxy-6,8,10,14-eicosatetraenoic acid; 5(S),12(R)-Dihydroxy-6(Z),8(e),10(e),14(Z)-eicosatetraenoate; 5(S),12(R)-Dihydroxy-6(Z),8(e),10(e),14(Z)-eicosatetraenoic acid; 5(S),12(R)-Dihydroxy-6(Z),8(e),10(e),14(Z)-icosatetraenoate; 5(S),12(R)-Dihydroxy-6(Z),8(e),10(e),14(Z)-icosatetraenoic acid; 5(S),12(R)-Dihydroxy-6-cis,8,10-trans,14-cis-eicosatetraenoic acid; 5,12 DiHETE; 5,12 HETE; 5,12-Dihete; 5,12-Dihydroxy-6,10-trans -8,14-cis -eicosatetraenoate; 5,12-Dihydroxy-6,10-trans -8,14-cis -eicosatetraenoic acid; 5,12-Hete; 5S,12R-Dihydroxy-6Z,8E,10E,14Z-eicosatetraenoate; 5S,12R-Dihydroxy-6Z,8E,10E,14Z-eicosatetraenoic acid; LTB4; Leucotriene b4; Leukotriene B4; Leukotriene b; Leukotriene b 4; Leukotriene b-4; Leukotriene b4 ethanol solution; Leukotrienes b; b-4, Leukotriene; cis-Leukotriene b4
|
| Function |
Leukotriene B4 is the major metabolite in neutrophil polymorphonuclear leukocytes. Leukotrienes are metabolites of arachidonic acid derived from the action of 5-LO (5-lipoxygenase). The immediate product of 5-LO is LTA4 (leukotriene A4), which is enzymatically converted into either LTB4 (leukotriene B4) by LTA4 hydrolase or LTC4 (leukotriene C4) by LTC4 synthase. The regulation of leukotriene production occurs at various levels, including expression of 5-LO, translocation of 5-LO to the perinuclear region, and phosphorylation to either enhance or inhibit the activity of 5-LO. Biologically active LTB4 is metabolized by omega-oxidation carried out by specific cytochrome P450s (CYP4F) followed by beta-oxidation from the omega-carboxy position and after CoA ester formation. Other specific pathways of leukotriene metabolism include the 12-hydroxydehydrogenase/15-oxo-prostaglandin-13-reductase that form a series of conjugated diene metabolites that have been observed to be excreted in human urine. Metabolism of LTC4 occurs by sequential peptide cleavage reactions involving a gamma-glutamyl transpeptidase that forms LTD4 (leukotriene D4) and a membrane-bound dipeptidase that converts LTD4 into LTE4 (leukotriene E4) before omega-oxidation. These metabolic transformations of the primary leukotrienes are critical for termination of their biological activity, and defects in expression of participating enzymes may be involved in specific genetic disease. The term leukotriene was coined to indicate the presence of three conjugated double bonds within the 20-carbon structure of arachidonic acid as well as the fact that these compounds were derived from leucocytes such as PMNNs or transformed mast cells. Interestingly, most of the cells known to express 5-LO are of myeloid origin, which includes neutrophils, eosinophils, mast cells, macrophages, basophils, and monocytes. Leukotriene biosynthesis begins with the specific oxidation of arachidonic acid by a free radical mechanism as a consequence of interaction with 5-LO. The first enzymatic step involves the abstraction of a hydrogen atom from C-7 of arachidonate followed by the addition of molecular oxygen to form 5-HpETE (5-hydroperoxyeicosatetraenoic acid). A second enzymatic step is also catalyzed by 5-LO and involves removal of a hydrogen atom from C-10, resulting in the formation of the conjugated triene epoxide LTA4. LTA4 must then be released by 5-LO and encounter either LTA4-H (LTA4 hydrolase) or LTC4-S [LTC4 (leukotriene C4) synthase]. LTA4-H can stereospecifically add water to C-12 while retaining a specific double-bond geometry, leading to LTB4 [leukotriene B4, 5(S),12(R)-dihydroxy-6,8,10,14-(Z,E,E,Z)-eicosatetraenoic acid]. If LTA4 encounters LTC4-S, then the reactive epoxide is opened at C-6 by the thiol anion of glutathione to form the product LTC4 [5(S)-hydroxy-6(R)-S-glutathyionyl-7,9,11,14- (E,E,Z,Z)-eicosatetraenoic acid], essentially a glutathionyl adduct of oxidized arachidonic acid. Both of these terminal leukotrienes are biologically active in that specific GPCRs recognize these chemical structures and receptor recognition initiates complex intracellular signalling cascades. In order for these molecules to serve as lipid mediators, however, they must be released from the biosynthetic cell into the extracellular milieu so that they can encounter the corresponding GPCRs. Surprising features of this cascade include the recognition of the assembly of critical enzymes at the perinuclear region of the cell and even localization of 5-LO within the nucleus of some cells. Under some situations, the budding phagosome has been found to assemble these proteins. Non-enzymatic proteins such as FLAP are now known as critical partners of this protein-machine assembly. An unexpected pathway of leukotriene biosynthesis involves the transfer of the chemically reactive intermediate, LTA4, from the biosynthetic cell followed by conversion into LTB4 or LTC4 by other cells that do not express 5-LO. Leukotrienes are eicosanoids. The eicosanoids consist of the prostaglandins (PGs), thromboxanes (TXs), leukotrienes (LTs), and lipoxins (LXs). The PGs and TXs are collectively identified as prostanoids. Prostaglandins were originally shown to be synthesized in the prostate gland, thromboxanes from platelets (thrombocytes), and leukotrienes from leukocytes, hence the derivation of their names. All mammalian cells except erythrocytes synthesize eicosanoids. These molecules are extremely potent, able to cause profound physiological effects at very dilute concentrations. All eicosanoids function locally at the site of synthesis, through receptor-mediated G-protein linked signalling pathways.
|
 click to show the details of this protein
click to show the details of this protein
 click to show the details of experiment for validating this pair
click to show the details of experiment for validating this pair