| General Information of MET (ID: META00940) |
| Name |
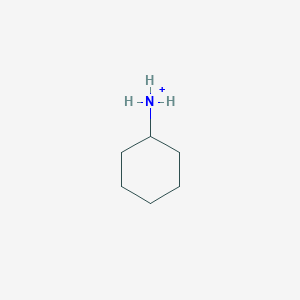
Cyclohexylammonium
|
| Synonyms |
Click to Show/Hide Synonyms of This Metabolite
CYCLOHEXYLAMMONIUM ion; Cyclohexylamine
|
| Source |
Drug
|
| Structure Type |
Cyclohexylamines (Click to Show/Hide the Complete Structure Type Hierarchy)
Organic nitrogen compounds
Organonitrogen compounds
Cyclohexylamines
|
| PubChem CID |
|
| HMDB ID |
|
| Formula |
C6H14N
|
| Structure |
<iframe style="width: 300px; height: 300px;" frameborder="0" src="https://embed.molview.org/v1/?mode=balls&cid=1549093"></iframe>
|
 |
|
3D MOL
|
2D MOL
|
|
Click to Show/Hide the Molecular/Functional Data (External Links/Property/Function) of This Metabolite
|
| DrugBank ID |
|
| ChEBI ID |
|
| Physicochemical Properties |
Molecular Weight |
100.18 |
Topological Polar Surface Area |
27.6 |
| XlogP |
1.5 |
Complexity |
46.1 |
| Heavy Atom Count |
7 |
Rotatable Bond Count |
N.A. |
| Hydrogen Bond Donor Count |
1 |
Hydrogen Bond Acceptor Count |
N.A. |
| Function |
Cyclohexylammonium is classified as a member of the Cyclohexylamines. Cyclohexylamines are organic compounds containing a cyclohexylamine moiety, which consist of a cyclohexane ring attached to an amine group. Cyclohexylammonium is considered to be practically insoluble (in water) and basic
|
|
Regulatory Network
|
|
|
|
|
|
|
|
|
 click to show the details of this protein
click to show the details of this protein
 click to show the details of experiment for validating this pair
click to show the details of experiment for validating this pair

