| General Information of MET (ID: META00891) |
| Name |
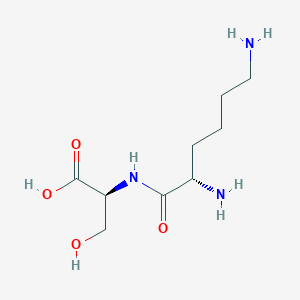
Lysylserine
|
| Synonyms |
Click to Show/Hide Synonyms of This Metabolite
K-S dipeptide; KS; KS dipeptide; L-Lys-L-ser; L-Lysyl-L-serine; Lys-Ser; Lysine serine dipeptide; Lysine-serine dipeptide; Lysyl-serine; Lysyl-serine dihydrochloride; Lysyl-serine, (D-lys-L-ser)-isomer; Lysyl-serine, bis(trifluoroacetate) salt; Lysyl-serine, mono-acetate salt; Lysylserine; N-L-Lysyl-L-serine; N-Lysylserine; T Cell modulatory peptide; TCMP-80
|
| Source |
Aliphatic acyclic compounds
|
| Structure Type |
Amino acids, peptides, and analogues (Click to Show/Hide the Complete Structure Type Hierarchy)
Organic acids and derivatives
Carboxylic acids and derivatives
Amino acids, peptides, and analogues
|
| PubChem CID |
|
| HMDB ID |
|
| Formula |
C9H19N3O4
|
| Structure |
<iframe style="width: 300px; height: 300px;" frameborder="0" src="https://embed.molview.org/v1/?mode=balls&cid=151428"></iframe>
|
 |
|
3D MOL
|
2D MOL
|
|
Click to Show/Hide the Molecular/Functional Data (External Links/Property/Function) of This Metabolite
|
| ChEBI ID |
|
| ChemSpider ID |
|
| Physicochemical Properties |
Molecular Weight |
233.27 |
Topological Polar Surface Area |
139 |
| XlogP |
-4.5 |
Complexity |
235 |
| Heavy Atom Count |
16 |
Rotatable Bond Count |
8 |
| Hydrogen Bond Donor Count |
5 |
Hydrogen Bond Acceptor Count |
6 |
| Function |
Lysylserine is a dipeptide composed of lysine and serine. It is an incomplete breakdown product of protein digestion or protein catabolism. Dipeptides are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. Some dipeptides are known to have physiological or cell-signalling effects although most are simply short-lived intermediates on their way to specific amino acid degradation pathways following further proteolysis.
|
|
Regulatory Network
|
|
|
|
|
|
|
|
|
 click to show the details of this protein
click to show the details of this protein
 click to show the details of experiment for validating this pair
click to show the details of experiment for validating this pair

