|
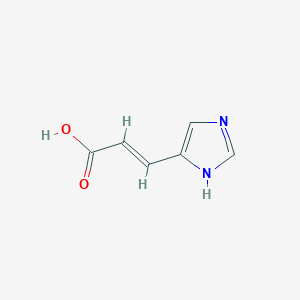
Urocanic acid (CAS: 104-98-3) is a breakdown (deamination) product of histidine. In the liver, urocanic acid is an intermediate in the conversion of histidine to glutamic acid, whereas, in the epidermis, it accumulates and may be both a UV protectant and an immunoregulator. Urocanic acid (UA) exists as a trans isomer (t-UA, approximately 30 mg/cm2) in the uppermost layer of the skin (stratum corneum). t-UA is formed as the cells of the second layer of the skin become metabolically inactive. During this process, proteins and membranes degrade, histidine is released, and histidase (histidine ammonia lyase) catalyzes the deamination of histidine to form t-UA. t-UA accumulates in the epidermis until removal by either the monthly skin renewal cycle or sweat. Upon absorption of UV light, the naturally occurring t-UA isomerizes to its cis form, c-UA. Because DNA lesions (e.g., pyrimidine dimers) in the lower epidermis can result from UV-B absorption, initial research proposed that t-UA acted as a natural sunscreen absorbing UV-B in the stratum corneum before the damaging rays could penetrate into lower epidermal zones. Researchers have found that c-UA also suppresses contact hypersensitivity and delayed hypersensitivity, reduces the Langerhans cell count in the epidermis, prolongs skin-graft survival time, and affects natural killer cell activity. (E)-Urocanic acid is found in mushrooms. It has been isolated from Coprinus atramentarius (common ink cap) and Phallus impudicus (common stinkhorn).
|
 click to show the details of this protein
click to show the details of this protein
 click to show the details of experiment for validating this pair
click to show the details of experiment for validating this pair